Metropolis-Hastings#
Integral of \(e^{-|x^3|}\)#
import numpy as np
import matplotlib.pyplot as plt
import scipy.integrate
seed = 239
rng = np.random.default_rng(seed)
def f(x):
return np.exp(-np.abs(x**3))
N = 10**4 # generate N samples
x = np.zeros(N)
x[0] = 0 # starting point
count = 0
for i in range(N-1):
# Proposal
x_try = x[i] + rng.normal(scale = 2)
# Accept the move or stay where we are
u = rng.uniform()
if u <= f(x_try) / f(x[i]):
x[i+1] = x_try
count = count + 1
else:
x[i+1] = x[i]
print("Acceptance fraction = %g" % (count/N,))
# Generate the same number of samples using the rejection method with uniform sampling
xmax = 4
xu = -xmax + 2*xmax*rng.uniform(size = 10*N) # Generate x values between +-xmax
y = rng.uniform(size = 10*N)
xu = xu[np.where(y <= f(xu))]
xu = xu[:count]
# Plot the distribution
counts, bins = np.histogram(x, bins=100, density = True)
plt.stairs(counts, bins, label = 'Metropolis')
counts, bins = np.histogram(xu, bins=100, density = True)
plt.stairs(counts, bins, label = 'Rejection method')
norm,_ = scipy.integrate.quad(f,-np.inf,np.inf)
xx = np.linspace(min(x),max(x),1000)
plt.plot(xx, f(xx)/norm, 'k:')
plt.yscale('log')
plt.legend()
plt.show()
# Plot the first 1000 samples to see the time series
plt.clf()
t = np.arange(N)
plt.plot(t[:1000], x[:1000])
plt.plot(t[:1000], xu[:1000] - 3.5)
plt.show()
# Plot the point-to-point correlation
plt.clf()
plt.figure(figsize=(4,7))
plt.plot(x[:-1], x[1:], '.')
plt.plot(xu[:-1], xu[1:] - 3.5, '.')
plt.show()
Acceptance fraction = 0.3596
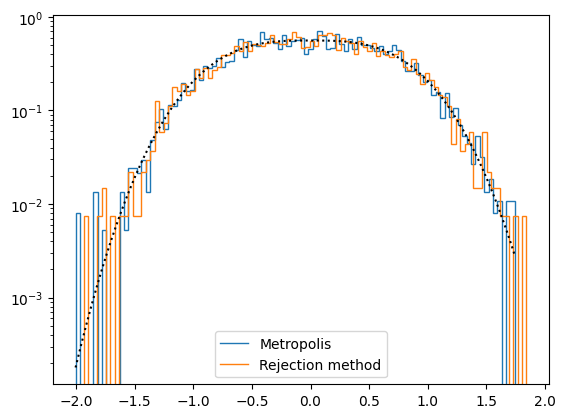
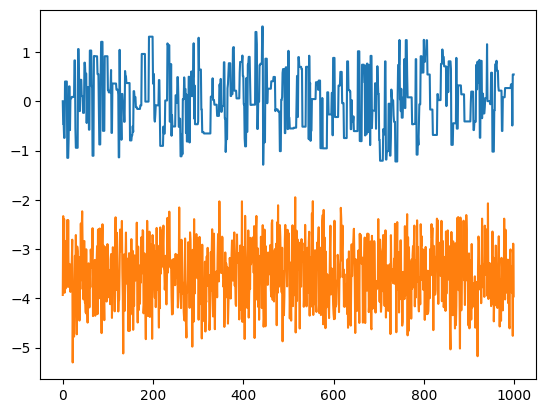
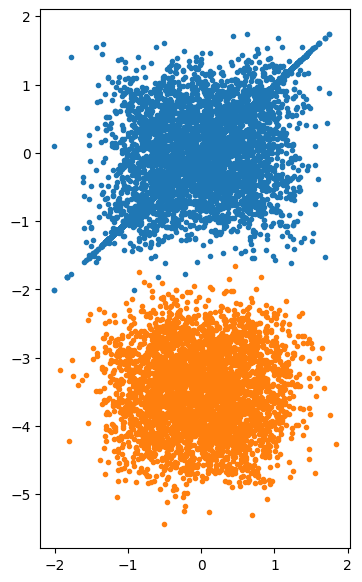
<Figure size 640x480 with 0 Axes>
Notes:
We started at \(x=0\) which is right in the middle of the high probability region. If instead you start at a point that has a low probability (e.g. try \(x=-5\) as initial condition in line 7), you will see that the chain has a “burn in” period in which it moves towards the higher probability regions. The part of the chain should be discarded.
The time series for the MCMC shows clear correlations, whereas the rejection method samples are uncorrelated. This can be seen in the plot of \(x_i\) vs \(x_{i+1}\) – in particular you can see the line of points where the proposed jump was not accepted.
Try changing the
scalein the normal distribution we’re using for the jump and see how the acceptance ratio and the correlations behave. You want to choose a jump size that is large enough to explore the probability distribution, but small enough to get a reasonable acceptance rate for the jumps. An acceptance rate of around \(0.2\)–\(0.3\) is good to aim for.
Exoplanet orbit#
def rv(t, P, x):
# Calculates the radial velocity of a star orbited by a planet
# at the times in the vector t
# extract the orbit parameters
# P, t and tp in days, mp in Jupiter masses, v0 in m/s
mp, e, omega, tp, v0 = x
# mean anomaly
M = 2*np.pi * (t-tp) / P
# velocity amplitude
K = 204 * P**(-1/3) * mp / np.sqrt(1.0-e*e) # m/s
# solve Kepler's equation for the eccentric anomaly E - e * np.sin(E) = M
# Iterative method from Heintz DW, "Double stars", Reidel, 1978
# first guess
E = M + e*np.sin(M) + ((e**2)*np.sin(2*M)/2)
while True:
E0 = E
M0 = E0 - e*np.sin(E0)
E = E0 + (M-M0)/(1.0 - e*np.cos(E0))
if np.max(np.abs((E-E0))) < 1e-6:
break
# evaluate the velocities
theta = 2.0 * np.arctan( np.sqrt((1+e)/(1-e)) * np.tan(E/2))
vel = v0 + K * ( np.cos(theta + omega) + e * np.cos(omega))
return vel
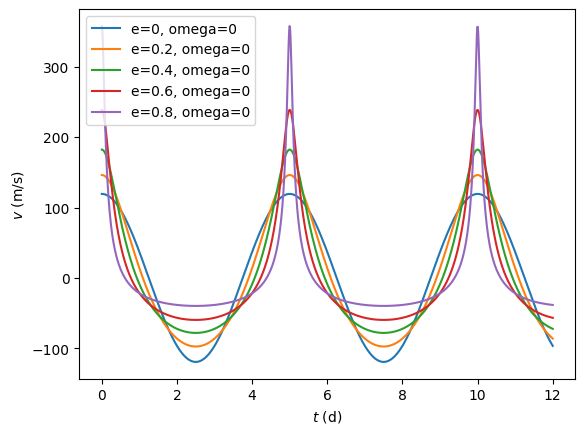
# Make some plots to show what the velocity curves look like
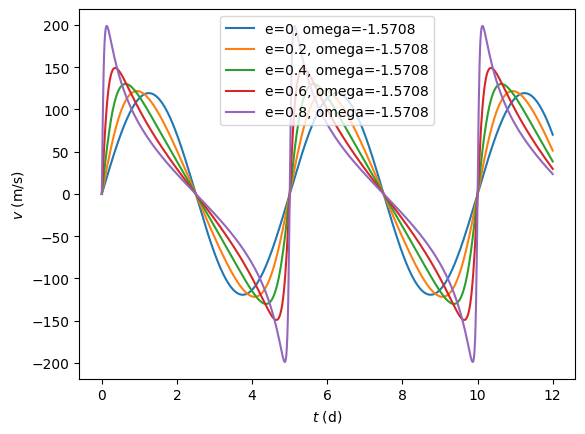
t = np.linspace(0,12,num=1000)
for e in np.arange(0.0,1.0,0.2):
v = rv(t, 5.0, [1.0, e, 0.0, 0.0, 0.0])
plt.plot(t,v, label='e=%g, omega=%g' % (e,0.0))
plt.ylabel(r'$v\ (\mathrm{m/s})$')
plt.xlabel(r'$t\ (\mathrm{d})$')
plt.legend()
plt.show()
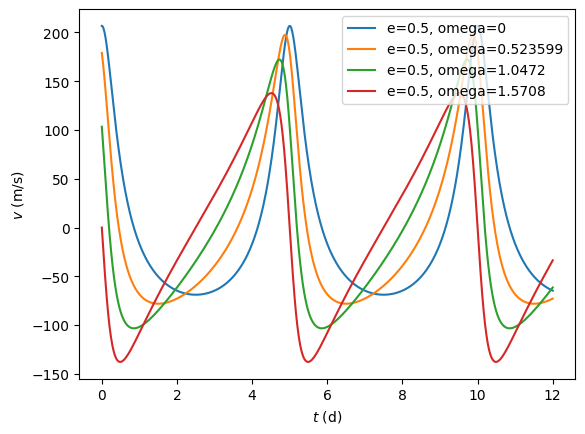
plt.clf()
for e in np.arange(0.0,1.0,0.2):
v = rv(t, 5.0, [1.0, e, -np.pi/2, 0.0, 0.0])
plt.plot(t,v, label='e=%g, omega=%g' % (e,-np.pi/2))
plt.ylabel(r'$v\ (\mathrm{m/s})$')
plt.xlabel(r'$t\ (\mathrm{d})$')
plt.legend()
plt.show()
plt.clf
for omega in np.linspace(0.0,np.pi/2,4):
v = rv(t, 5.0, [1.0, 0.5, omega, 0.0, 0.0])
plt.plot(t,v, label='e=%g, omega=%g' % (0.5, omega))
plt.ylabel(r'$v\ (\mathrm{m/s})$')
plt.xlabel(r'$t\ (\mathrm{d})$')
plt.legend()
plt.show()
def f(x, tobs, vobs, eobs):
chisq = np.sum(((vobs-rv(tobs, P, x))/eobs)**2)
return -chisq/2
# Observations
# These are for HD145675 from Butler et al. 2003
tobs, vobs, eobs = np.loadtxt('rvs.txt', unpack=True)
# Number of samples to generate
N = 10**5
x = np.zeros((N, 5))
# initial guess
# P, mp, e, omega, tp, v0
P = 1724
x[0] = [1.0, 0.0, 0.0, 0.0, 0.0]
# and the widths for the jumps
widths = (0.03, 0.03, 0.03, 3.0, 1.0)
count = 0
for i in range(N-1):
# Proposal
ii = np.random.randint(0, 5)
x_try = np.copy(x[i])
x_try[ii] += rng.normal(scale = widths[ii])
#x_try[2] = (x_try[2]) % 1 # keep e between zero and 1
# Accept the move or stay where we are
u = rng.uniform()
if u <= np.exp(f(x_try, tobs,vobs,eobs) - f(x[i], tobs,vobs,eobs)):
x[i+1] = np.copy(x_try)
count = count + 1
else:
x[i+1] = np.copy(x[i])
print("Acceptance fraction = %g" % (count/N,))
Acceptance fraction = 0.39833
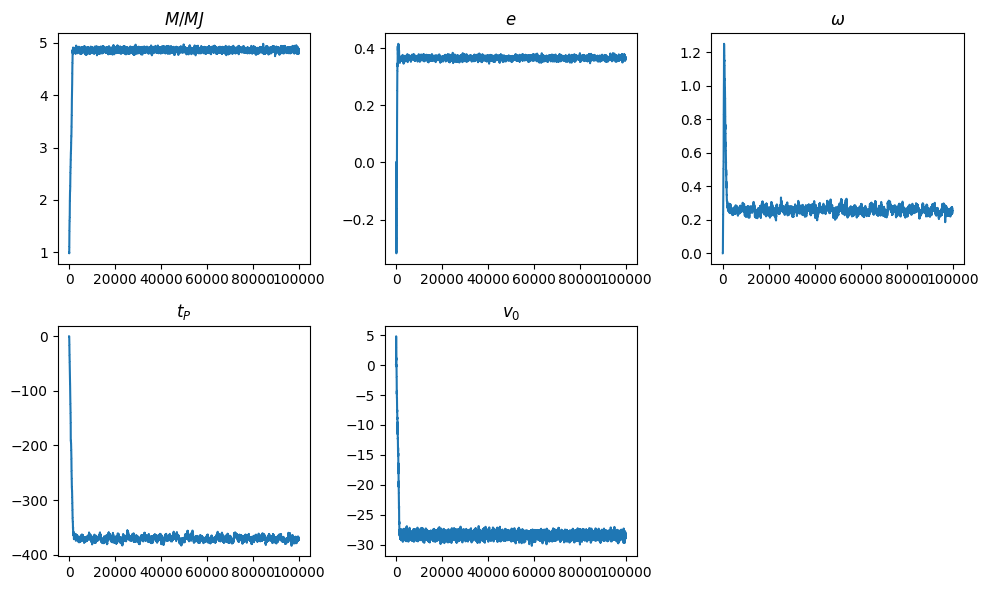
plt.figure(figsize=(10,6))
titles = (r'$M/MJ$', r'$e$', r'$\omega$', r'$t_P$', r'$v_0$')
for i, title in enumerate(titles):
plt.subplot(2,3,i+1)
plt.title(title)
plt.plot(list(range(N)), x[:, i])
plt.tight_layout()
plt.show()
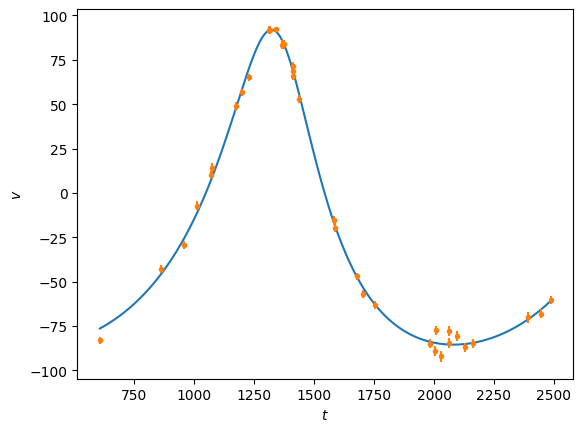
plt.clf()
t = np.linspace(tobs[0], tobs[-1], 1000)
plt.plot(t, rv(t,P, x[-1]), 'C0')
plt.plot(tobs, vobs, 'C1.')
plt.errorbar(tobs, vobs, eobs, fmt='none', ecolor='C1')
plt.ylabel(r'$v$')
plt.xlabel(r'$t$')
plt.show()
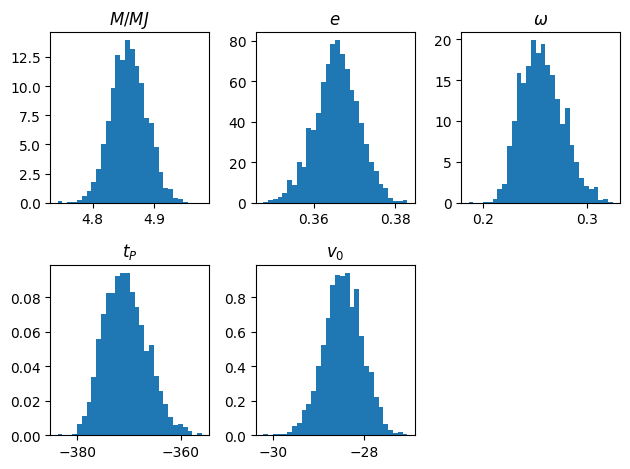
# Reject the burn in phase
x1 = x[int(0.5*N):]
plt.clf()
for i, title in enumerate(titles):
plt.subplot(2,3,i+1)
plt.title(title)
plt.hist(x1[:, i], density=True, bins=30)
plt.tight_layout()
plt.show()
# https://corner.readthedocs.io/en/latest/
import corner
figure = corner.corner(x1,
labels = [r'$M/M_J$', r'$e$', r'$\omega$', r'$t_P$', r'$v_0$'],
show_titles=True)
---------------------------------------------------------------------------
ModuleNotFoundError Traceback (most recent call last)
Cell In[7], line 2
1 # https://corner.readthedocs.io/en/latest/
----> 2 import corner
4 figure = corner.corner(x1,
5 labels = [r'$M/M_J$', r'$e$', r'$\omega$', r'$t_P$', r'$v_0$'],
6 show_titles=True)
ModuleNotFoundError: No module named 'corner'
