Homework 6 solutions#
Harmonic oscillator Hamiltonian#
(a) First check what happens to the Hamiltonian and pseudo-Hamiltonian when we make an update:
import numpy as np
import matplotlib.pyplot as plt
import scipy.linalg
import sympy.matrices
from sympy.printing import pprint
# declare the step size h as a symbol
h = sympy.Symbol('h')
def J_leapfrog(h):
a = sympy.matrices.Matrix([[1,0],[-h/2,1]])
b = sympy.matrices.Matrix([[1,h],[0,1]])
return a*b*a
x = sympy.Symbol('x')
v = sympy.Symbol('v')
xv = sympy.matrices.Matrix([x, v])
print("Start with")
pprint(xv)
xvnew = J_leapfrog(h) * xv
xnew = xvnew[0]
vnew = xvnew[1]
print("\nTime evolve changes this to ")
pprint(xvnew)
H = x**2 + v**2
Hnew = xnew**2 + vnew**2
print("\nDelta H = ")
pprint(sympy.simplify(Hnew-H))
Htilde = x**2 + v**2/(1-h**2/4)
Htildenew = xnew**2 + vnew**2/(1-h**2/4)
print("\nDelta Htilde = ")
pprint(sympy.simplify(Htildenew-Htilde))
Start with
⎡x⎤
⎢ ⎥
⎣v⎦
Time evolve changes this to
⎡ ⎛ 2⎞ ⎤
⎢ ⎜ h ⎟ ⎥
⎢ h⋅v + x⋅⎜1 - ──⎟ ⎥
⎢ ⎝ 2 ⎠ ⎥
⎢ ⎥
⎢ ⎛ ⎛ 2⎞ ⎞⎥
⎢ ⎜ ⎜ h ⎟ ⎟⎥
⎢ ⎛ 2⎞ ⎜ h⋅⎜1 - ──⎟ ⎟⎥
⎢ ⎜ h ⎟ ⎜ ⎝ 2 ⎠ h⎟⎥
⎢v⋅⎜1 - ──⎟ + x⋅⎜- ────────── - ─⎟⎥
⎣ ⎝ 2 ⎠ ⎝ 2 2⎠⎦
Delta H =
3 ⎛ 3 2 2 2 2 ⎞
h ⋅⎝h ⋅x - 4⋅h ⋅v⋅x + 4⋅h⋅v - 4⋅h⋅x + 8⋅v⋅x⎠
───────────────────────────────────────────────
16
Delta Htilde =
0
This shows that whereas the Hamiltonian is not conserved (\(\Delta H\neq 0\)), the pseudo Hamiltonian is (\(\Delta \tilde{H}=0\)).
(b) Now implement leapfrog for the harmonic oscillator
def leapfrog(nsteps, dt, x0, v0):
x = np.zeros(nsteps+1)
v = np.zeros(nsteps+1)
x[0] = x0
v[0] = v0
for i in range(1, nsteps+1):
a = -x[i-1]
v12 = v[i-1] + a * dt/2
x[i] = x[i-1] + v12 * dt
a = -x[i]
v[i] = v12 + a * dt/2
return x, v
nsteps = 100
dt = 2*np.pi / nsteps
t = np.arange(nsteps+1)*dt
x, v = leapfrog(nsteps, dt, 0, 1)
E = x**2 + v**2
E1 = x**2 + v**2 / (1-dt**2/4)
print(nsteps, np.max(np.abs(E-1)), E1[-1]-1)
fig = plt.figure(figsize=(8,2.5))
plt.subplot(131)
plt.plot(x, v)
plt.xlabel(r'$x$')
plt.ylabel(r'$x$')
plt.subplot(132)
plt.plot(t,x)
plt.plot(t,v)
plt.xlabel(r'$t$')
plt.ylabel(r'$x, v$')
plt.subplot(133)
plt.plot(t,E-1)
plt.plot(t,E1-1)
plt.xlabel(r'$t$')
plt.ylabel(r'E-1')
plt.tight_layout()
plt.show()
100 0.0009879354273425456 0.0009879354933575168
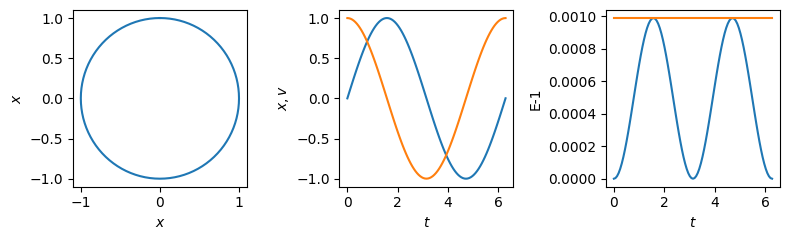
The plot on the right shows that whereas the energy oscillates up and down over a cycle, the pseudo-Hamiltonian remains constant as expected.
(c) Look at the scaling: (the original wording of the question had a typo, we need to look at the scaling for 1/2 and 1/4 of a cycle not 1 and 1/2 cycles)
nsteps = 10**np.linspace(1,6,6, dtype = 'int')
Evec = np.array([])
E1vec = np.array([])
for n in nsteps:
dt = 0.5 * np.pi / n
x, v = leapfrog(n, dt, 0, 1)
E = x**2 + v**2
x, v = leapfrog(n, dt, x[-1], v[-1])
E1 = x**2 + v**2
Evec = np.append(Evec, abs(E[-1]-1))
E1vec = np.append(E1vec, abs(E1[-1]-1))
plt.plot(nsteps, E1vec, label="1/2 cycle")
plt.plot(nsteps, Evec, label="1/4 cycle")
plt.plot(nsteps, 1/nsteps**2,':')
plt.plot(nsteps, 1/nsteps**6,':')
plt.plot(nsteps, 1e-16*nsteps**0.5,':')
plt.ylim((1e-16, 1))
plt.legend()
plt.yscale('log')
plt.xscale('log')
plt.show()
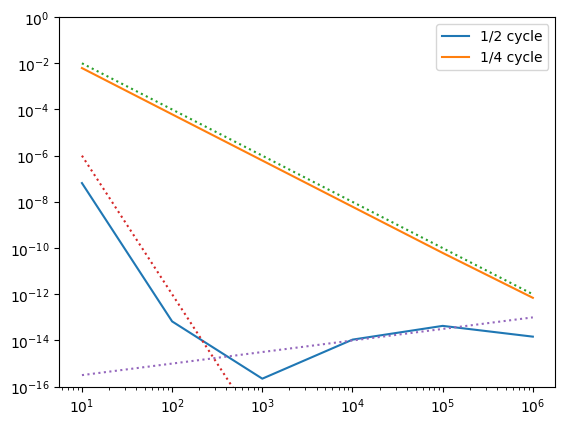
For 1/2 a cycle, the energy error is \(\propto 1/N^6\), whereas for 1/4 of a cycle we find \(\propto 1/N^2\). In our expression for \(\Delta H\) above, averaging over an orbit will leave only the \(h^6\) term, so the \(N^6\) scaling makes sense. Over 1/4 of an orbit, we don’t get the averaging effect. The largest error term is \(\propto h^3\) (local error), which then accumulates over the integration (global error) to give an \(N^2\) scaling.
Eigenvalue problem for the wave on a string#
Write the difference equation as
which is of the form
We can solve this using scipy.linalg.eigh. We take advantage of the ability of scipy.linalg.eigh to solve a “generalized eigenvalue problem” so we can pass it the \(\mathbf{b}\) vector directly.
n = 100
x = np.linspace(0,1,n)
h = x[1]-x[0] # grid spacing
h2 = h**2
#rho = np.ones(n)
rho = 1 + 10*x**2
A = np.diag(2*np.ones(n), k=0) + np.diag(-np.ones(n-1),k=-1) + np.diag(-np.ones(n-1),k=1)
w2, v = scipy.linalg.eigh(A, b=np.diag(rho))
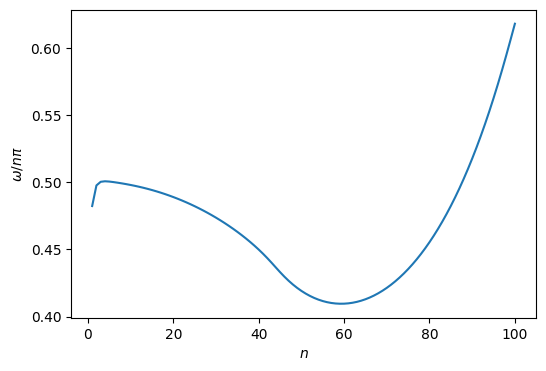
w2 = w2 / h**2
w=np.sqrt(w2)/np.pi
i = np.arange(len(w)) + 1
plt.figure(figsize=(6,4))
plt.plot(i, w/i)
plt.ylabel(r'$\omega/n\pi$')
plt.xlabel(r'$n$')
plt.show()
plt.clf()
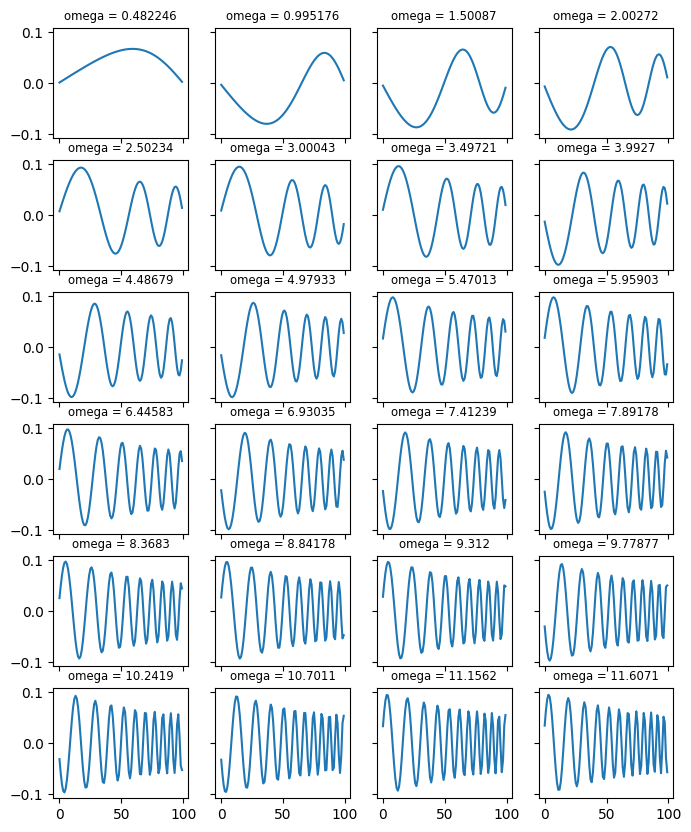
# Note that the n-th eigenvector is given by v[:, n-1] (not v[n-1]!)
fig, ax = plt.subplots(6, 4, sharex='all', sharey='all', figsize=(8,10))
for i in range(24):
ax[i//4, i%4].plot(np.arange(len(v[:,i])), v[:,i])
ax[i//4, i%4].set_title("omega = %lg" % (w[i]), fontsize='small')
<Figure size 640x480 with 0 Axes>
Electromagnetic wave#
ngrid = 1001
x = np.arange(ngrid)
# refractive indices
n1 = 1.0
n2 = 1.5
# alpha = dt/dx
alpha = 1.0/ max(n1,n2)
# initial wavepacket
E0 = np.ones(ngrid)
#E0 = np.sin(2.0 * np.pi * x / 20)
E0 = E0 * np.exp(-(x-ngrid//6)**2/800)
# refractive index array
n = np.ones(ngrid) # set all n = 1
n[ngrid//2:] = n2
n[:ngrid//2] = n1
# set the B field to be nE so the wave propagates to the right
# (-nE will go left)
B0 = E0 * n
# arrays for E and B
Ep = np.copy(E0)
Bp = np.copy(B0)
# do a first order step
E = Ep - alpha * (np.roll(Bp,-1) - np.roll(Bp,1)) / 2.0 / n**2
B = Bp - alpha * (np.roll(E,-1) - np.roll(E,1)) / 2.0
Enew = np.copy(E)
Bnew = np.copy(B)
# number of iterations
niter = int(0.7 * ngrid / alpha)
# set up plots and plot the starting profile
%matplotlib
plt.ion()
plt.plot(x,E,"C0:")
plt.plot(x,B,"C1:")
plt.plot([ngrid//2,ngrid//2],[-1.5,1.5], 'k', alpha=0.2, lw=2)
dat, = plt.plot(x, E, 'C0')
dat2, = plt.plot(x, B, 'C1')
plt.ylim((-1.5,1.5))
plt.xlim((x[0],x[-1]))
plt.show()
for k in range(niter):
Enew = Ep - alpha * (np.roll(B,-1) - np.roll(B,1)) / n**2
Bnew = Bp - alpha * (np.roll(E,-1) - np.roll(E,1))
# draw the updated data
dat.set_ydata(Enew)
dat2.set_ydata(Bnew)
plt.draw()
plt.pause(1e-3)
Ep[:] = E[:]
E[:] = Enew[:]
Bp[:] = B[:]
B[:] = Bnew[:]
plt.ioff()
plt.close()
%matplotlib inline
plt.plot(x, E, label='E')
plt.plot(x, B, label='B')
plt.ylim((-1.5,1.5))
plt.xlim((x[0],x[-1]))
# plot the analytic reflection and transmission coeffs
r = abs((n2-n1)/(n2+n1))
t = 2 * n1 / (n2+n1)
plt.plot([0,1000],[r,r],'k-.')
plt.plot([0,1000],[t,t],'k--')
plt.plot([0,1000],[-r,-r],'k-.')
plt.plot([0,1000],[-t,-t],'k--')
plt.legend()
plt.show()
Using matplotlib backend: <object object at 0x105bcf3c0>
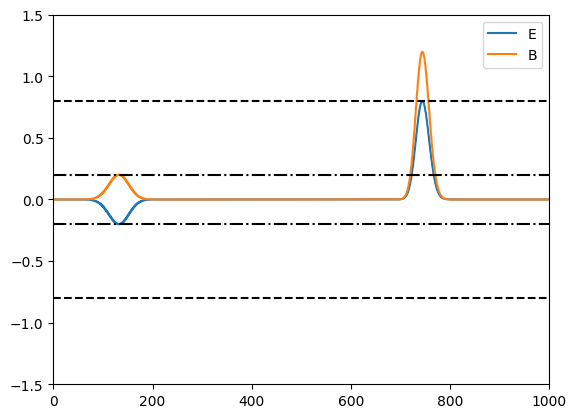
The dot-dashed and dashed lines show the expected amplitudes of the reflected and transmitted waves.
