Homework 4 solutions#
import numpy as np
import matplotlib.pyplot as plt
# Visualize matrices as a color map
def plot_matrices(A,titles=[]):
n = len(A)
if titles==[]:
titles = [""]*n
if n>4:
nx = 4
else:
nx = n
for j in range(int(np.floor(n/4))+1):
plt.clf()
plt.figure(figsize=(nx*4,4))
jmax = 4*(j+1)
if jmax > n:
jmax = n
for i,AA in enumerate(A[4*j:jmax]):
plt.subplot(1, nx, i+1)
plt.imshow(AA)
plt.colorbar()
plt.title(titles[4*j + i])
plt.show()
1. More on SVD#
# Visualize matrices as a color map
def plot_matrices(A,titles=[]):
n = len(A)
if titles==[]:
titles = [""]*n
if n>4:
nx = 4
else:
nx = n
for j in range(int(np.floor(n/4))+1):
plt.clf()
plt.figure(figsize=(nx*4,4))
jmax = 4*(j+1)
if jmax > n:
jmax = n
for i,AA in enumerate(A[4*j:jmax]):
plt.subplot(1, nx, i+1)
plt.imshow(AA)
plt.colorbar()
plt.title(titles[4*j + i])
plt.show()
m=10; n=6
A = np.random.rand(m*n).reshape(m,n)
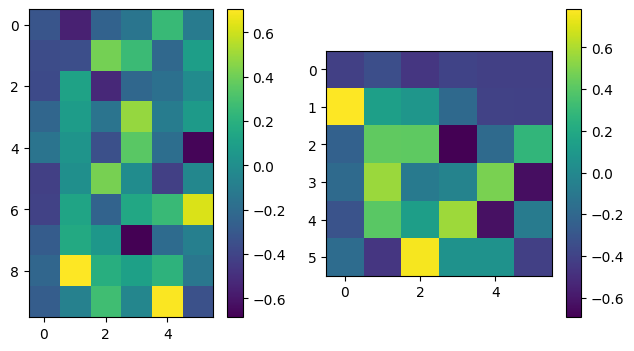
U, Sdiag, VT = np.linalg.svd(A, full_matrices=False)
plot_matrices([U,VT])
print("Singular values are: ", Sdiag)
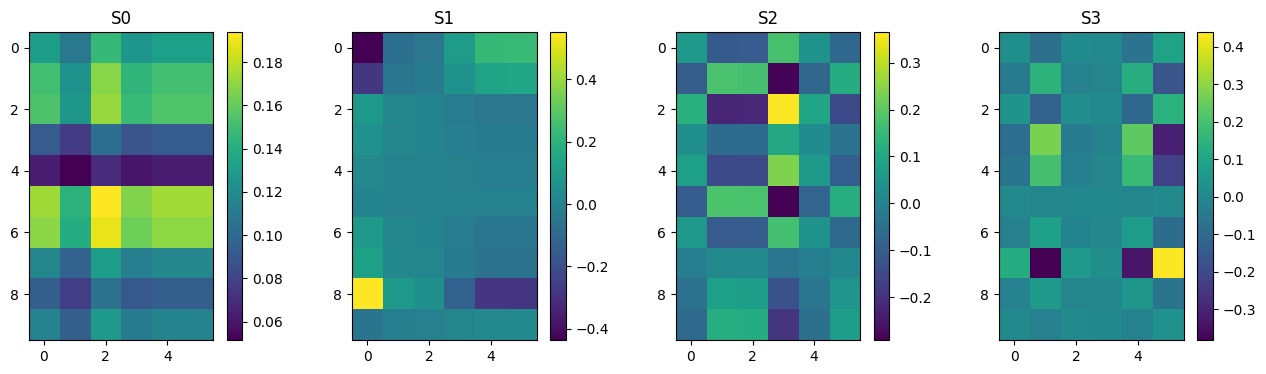
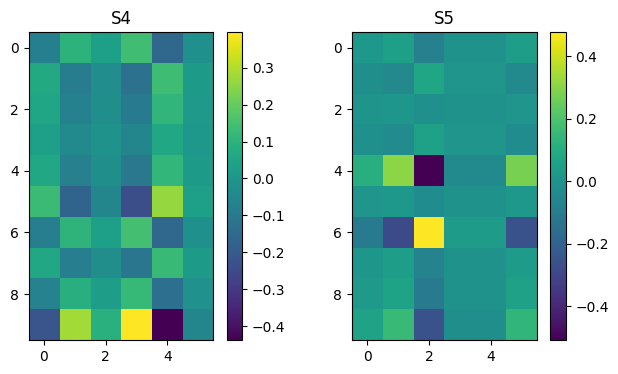
Ak = np.zeros((n, m, n))
for k in range(n):
Ak[k] = np.outer(U[:,k], VT[k,:])
plot_matrices(Ak[:],["S%d" % (i,) for i in range(n)])
Ar = np.zeros_like(A)
for k in range(n):
Ar = Ar + Sdiag[k] * Ak[k]
plot_matrices([A, Ar, A-Ar])
<Figure size 640x480 with 0 Axes>
Singular values are: [4.38084506 1.18504164 0.99288971 0.71181482 0.57488263 0.16930277]
<Figure size 640x480 with 0 Axes>
<Figure size 640x480 with 0 Axes>
<Figure size 640x480 with 0 Axes>
from PIL import Image
img = Image.open('github_logo.png')
A = np.asarray(img)[:,:,1]
plot_matrices([A])
m,n = np.shape(A)
print('shape = ', m,n)
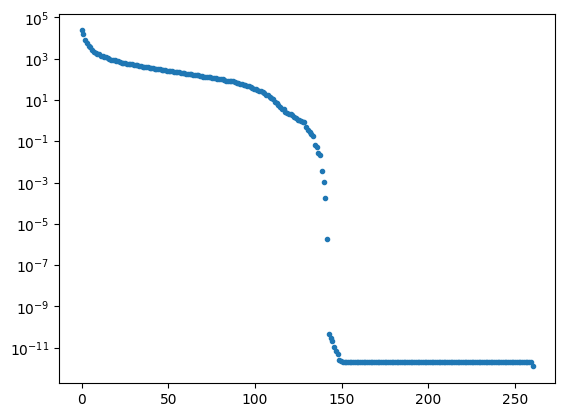
U, Sdiag, VT = np.linalg.svd(A, full_matrices=False)
nk = len(Sdiag)
plt.clf()
plt.plot(np.linspace(0,nk,nk),Sdiag,'.')
plt.yscale('log')
plt.show()
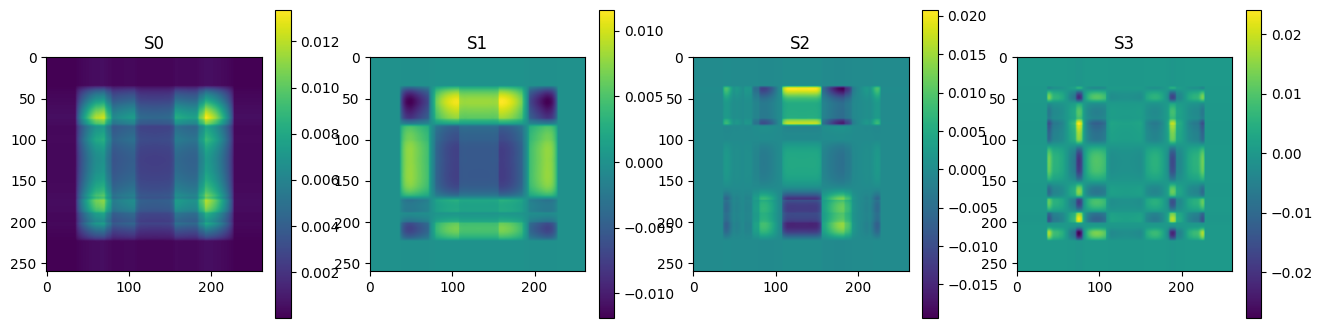
Ak = np.zeros((n, m, n))
for k in range(nk):
Ak[k] = np.outer(U[:,k], VT[k,:])
# plot some of the matrices
plot_matrices(Ak[:12],["S%d" % (i,) for i in range(n)])
<Figure size 640x480 with 0 Axes>
shape = 260 262
<Figure size 640x480 with 0 Axes>
<Figure size 640x480 with 0 Axes>
<Figure size 640x480 with 0 Axes>
<Figure size 640x480 with 0 Axes>
<Figure size 1600x400 with 0 Axes>
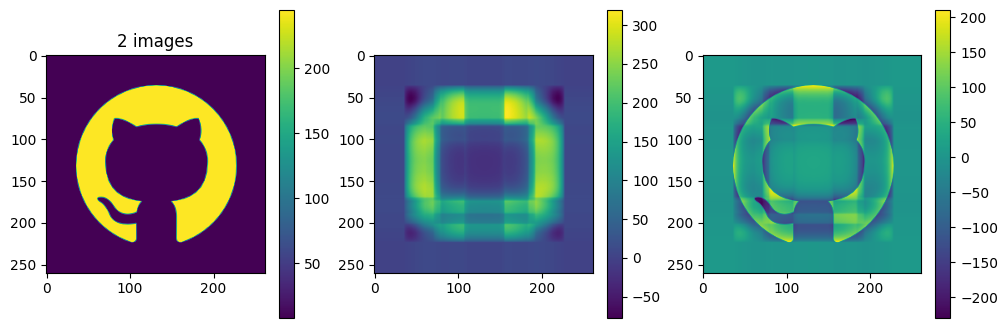
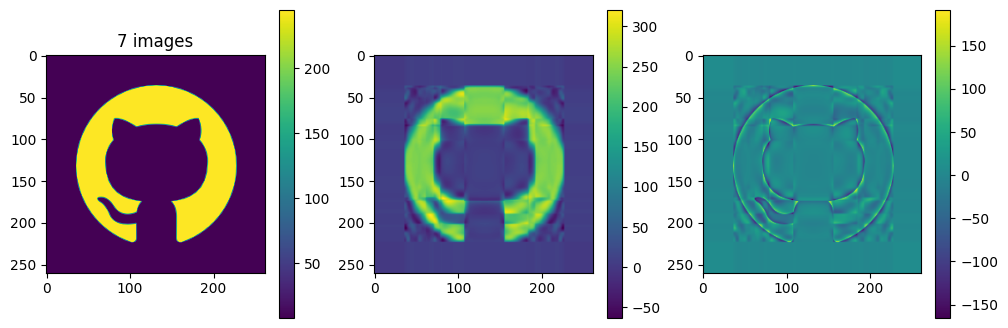
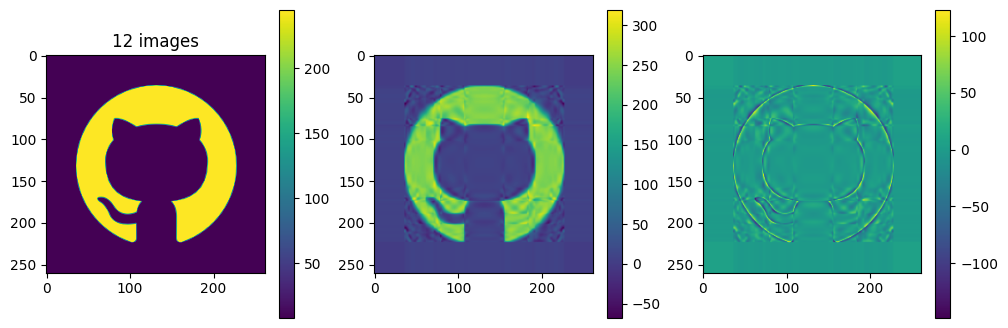
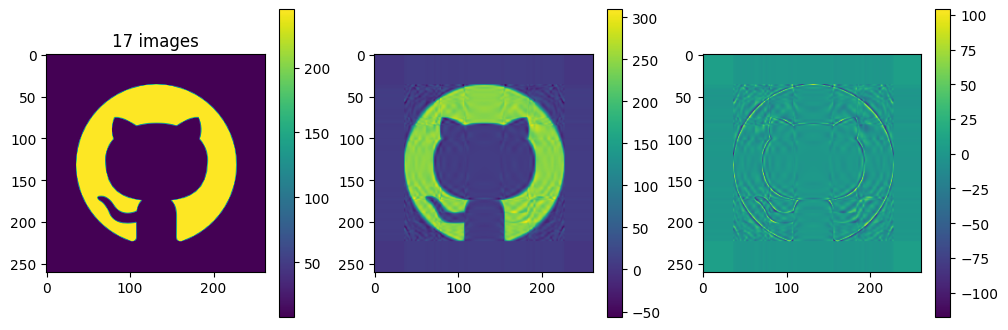
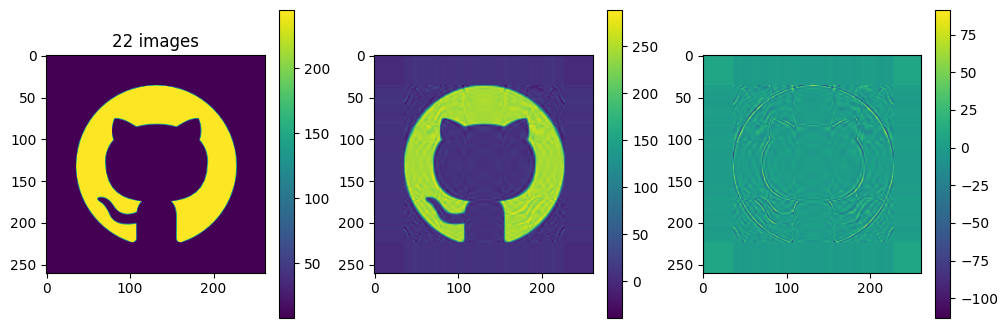
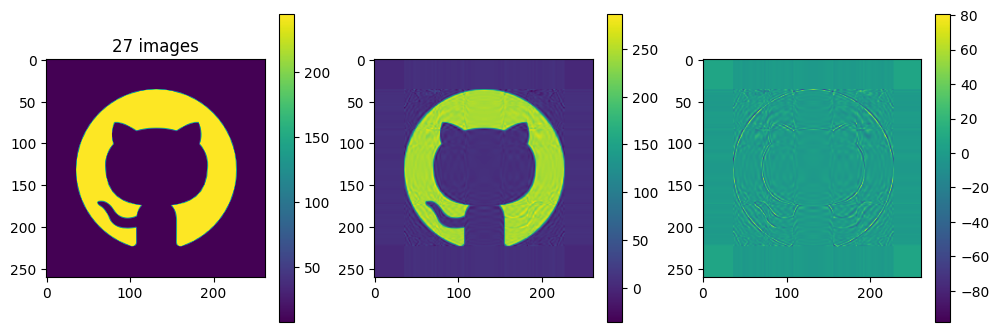
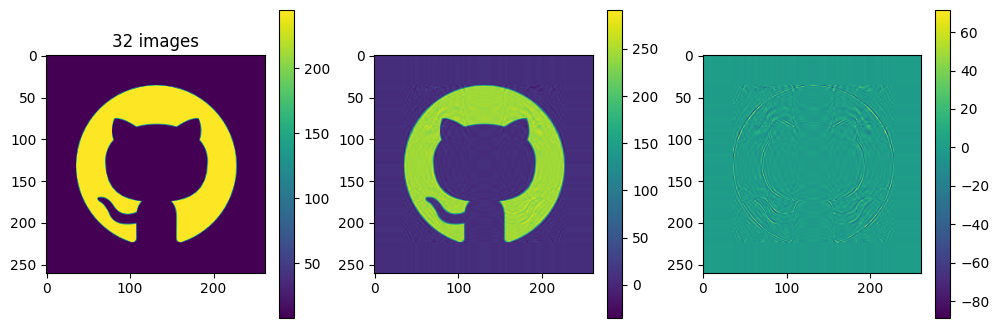
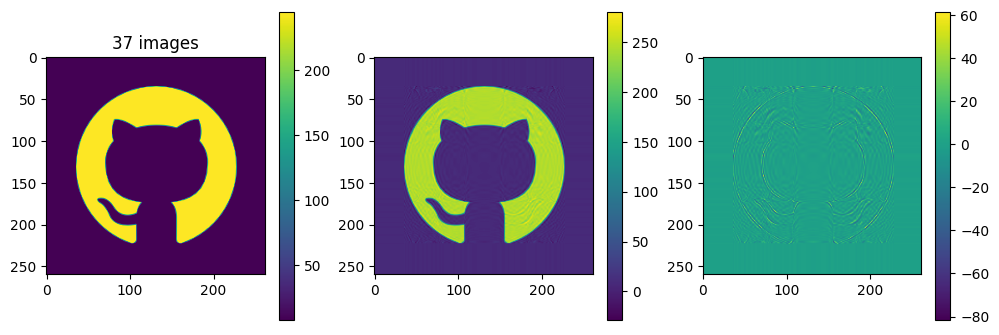
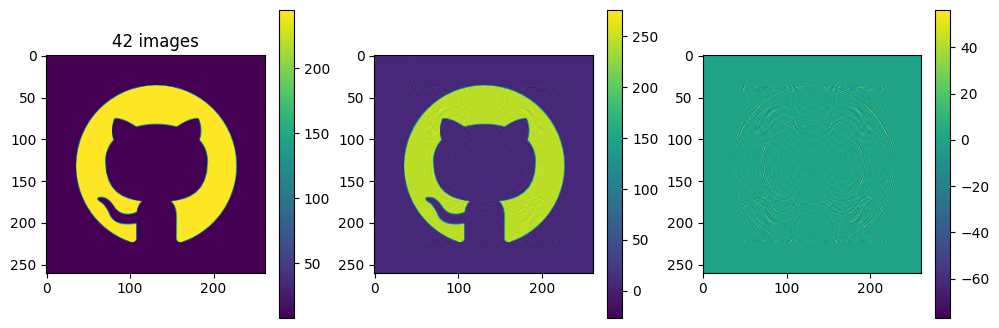
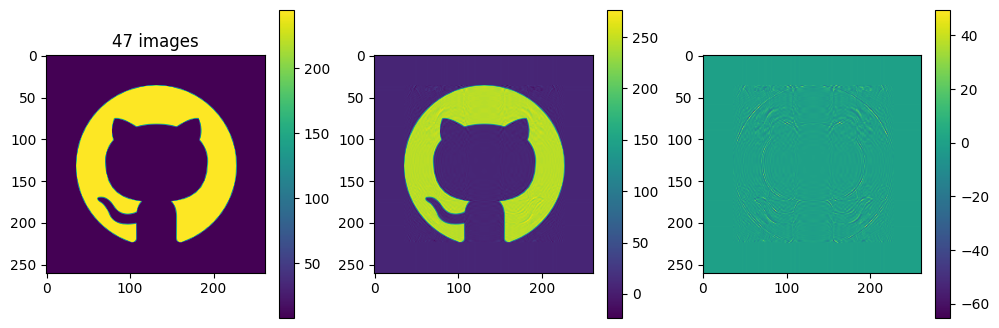
# now show the reconstructed images with different numbers of input matrices
for kmax in range(2,61,5):
Ar = np.zeros_like(A)
for k in range(kmax):
Ar = Ar + Sdiag[k] * Ak[k]
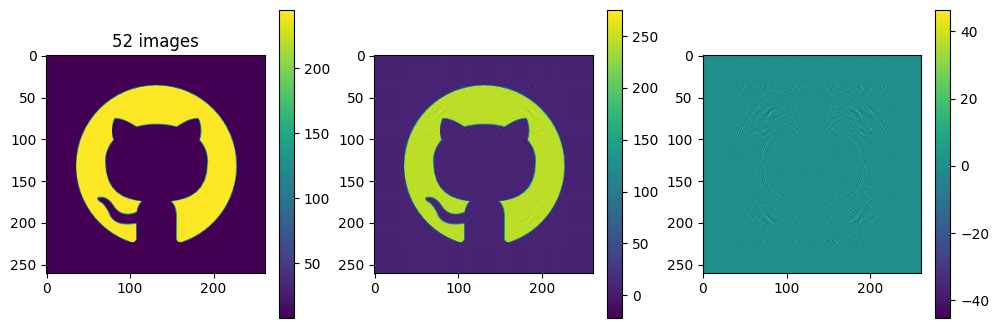
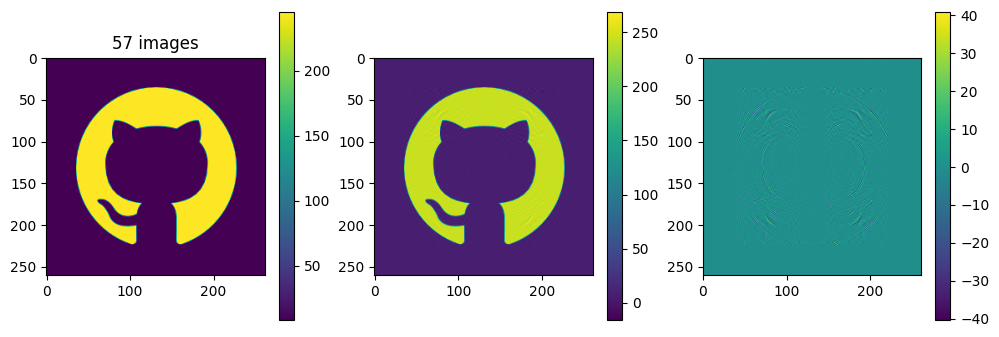
plot_matrices([A, Ar, A-Ar], titles=["%d images" % (kmax,), "", ""])
<Figure size 640x480 with 0 Axes>
<Figure size 640x480 with 0 Axes>
<Figure size 640x480 with 0 Axes>
<Figure size 640x480 with 0 Axes>
<Figure size 640x480 with 0 Axes>
<Figure size 640x480 with 0 Axes>
<Figure size 640x480 with 0 Axes>
<Figure size 640x480 with 0 Axes>
<Figure size 640x480 with 0 Axes>
<Figure size 640x480 with 0 Axes>
<Figure size 640x480 with 0 Axes>
<Figure size 640x480 with 0 Axes>
print(len(U[:,0]), len(VT[0,:]), nk)
260 262 260
print(20 * (1+260+262), 260*262, (262*260)/(20 * (1+260+262)))
10460 68120 6.512428298279159
Each of the component matrices is constructed from two vectors of length \(m\) and \(n\), so the total storage is \((m+n)\) for those vectors. If we keep \(N\) component matrices, we need \(N\) of these vectors and also \(N\) singular values, ie. a total of \(N(m+n+1)\) numbers. This should be compared to \(m\times n\) for the original matrix.
So in this example, it looks like about 20 components gives a reasonable approximation of the image – so we need \(20\times (1+260+262)=10460\) numbers.
The compression factor is therefore \((260\times 262)/10460\approx 6.5\) compared to storing the full \(m\times n\) matrix.
2. Fitting planetary orbits#
import numpy as np
import matplotlib.pyplot as plt
def rv(t, P, a):
# Calculates the radial velocity of a star orbited by a planet
# at the times in the vector t
# extract the orbit parameters
# P, t and tp in days, mp in Jupiter masses, v0 in m/s
mp, e, omega, tp, v0 = a
# mean anomaly
M = 2*np.pi * (t-tp) / P
# velocity amplitude
K = 204 * P**(-1/3) * mp / np.sqrt(1.0-e*e) # m/s
# solve Kepler's equation for the eccentric anomaly E - e * np.sin(E) = M
# Iterative method from Heintz DW, "Double stars", Reidel, 1978
# first guess
E = M + e*np.sin(M) + ((e**2)*np.sin(2*M)/2)
while True:
E0 = E
M0 = E0 - e*np.sin(E0)
E = E0 + (M-M0)/(1.0 - e*np.cos(E0))
if np.max(np.abs((E-E0))) < 1e-6:
break
# evaluate the velocities
theta = 2.0 * np.arctan( np.sqrt((1+e)/(1-e)) * np.tan(E/2))
vel = v0 + K * ( np.cos(theta + omega) + e * np.cos(omega))
return vel
def rv_model(t, P, a):
v = rv(t, P, a)
A = np.zeros([len(t), len(a)])
eps = 1e-8
b = np.copy(a) * (1 + eps)
v0 = rv(t, P, np.array([b[0],a[1],a[2],a[3],a[4]]))
v1 = rv(t, P, np.array([a[0],b[1],a[2],a[3],a[4]]))
v2 = rv(t, P, np.array([a[0],a[1],b[2],a[3],a[4]]))
v3 = rv(t, P, np.array([a[0],a[1],a[2],b[3],a[4]]))
v4 = rv(t, P, np.array([a[0],a[1],a[2],a[3],b[4]]))
A[:, 0] = (v0 - v) / (b[0]-a[0])
A[:, 1] = (v1 - v) / (b[1]-a[1])
A[:, 2] = (v2 - v) / (b[2]-a[2])
A[:, 3] = (v3 - v) / (b[3]-a[3])
A[:, 4] = (v4 - v) / (b[4]-a[4])
return v, A
# Observations
# These are for HD145675 from Butler et al. 2003
tobs, vobs, eobs = np.loadtxt('rvs.txt', unpack=True)
# initial guess
# P, mp, e, omega, tp, v0
P = 1724
a0 = np.array([1.0, 1e-3, 1e-3, 1e-3, 1e-3])
a = a0
err = 1.0
lam = 1e-3
while err > 1e-6:
v, A = rv_model(tobs, P, a)
A[:, 0] = A[:, 0] / eobs
A[:, 1] = A[:, 1] / eobs
A[:, 2] = A[:, 2] / eobs
A[:, 3] = A[:, 3] / eobs
A[:, 4] = A[:, 4] / eobs
r = (v-vobs)/eobs
lhs = A.T@A
lhs = lhs@(np.identity(len(a))*(1+lam))
rhs = -A.T@r
da = np.linalg.inv(lhs)@rhs
# artificially reduce the step to not overshoot e=1
a1 = a + 0.2*da
#print(a1, a)
v1, A1 = rv_model(tobs, P, a1)
A1[:, 0] = A1[:, 0] / eobs
A1[:, 1] = A1[:, 1] / eobs
A1[:, 2] = A1[:, 2] / eobs
A1[:, 3] = A1[:, 3] / eobs
A1[:, 4] = A1[:, 4] / eobs
r1 = (v1-vobs)/eobs
err = np.sum(r**2)
err1 = np.sum(r1**2)
print("err = %lg, a=(%lg, %lg, %lg, %lg, %lg), lam = %lg" % (err1 ,a1[0], a1[1], a1[2], a1[3], a1[4], lam))
if err1 > err:
lam = lam * 10
else:
lam = lam / 10
a = a1
if (err-err1) < 0.1:
break
print("Best-fit parameters: M = %lg, e = %lg, omega = %lg, tP = %lg, v0 = %lg " %
(a[0], a[1], a[2] % np.pi, a[3], a[4]))
err = 27681.8, a=(0.782451, -0.369395, 1.25645, 83.4323, -5.54384), lam = 0.001
err = 20054.1, a=(1.41799, -0.0958655, 3.1723, 397.376, -9.24603), lam = 0.0001
err = 14422.2, a=(2.07504, -0.259971, 4.42914, 762.861, -12.9742), lam = 1e-05
err = 9761.64, a=(2.58825, -0.194585, 3.86685, 619.679, -15.7538), lam = 1e-06
err = 6274.18, a=(3.0306, -0.236762, 3.5061, 522.647, -18.2403), lam = 1e-07
err = 4006.37, a=(3.39172, -0.277748, 3.43951, 504.4, -20.2629), lam = 1e-08
err = 2573.44, a=(3.68319, -0.303122, 3.41861, 498.425, -21.8904), lam = 1e-09
err = 1664.62, a=(3.91746, -0.319699, 3.40947, 495.657, -23.1969), lam = 1e-10
err = 1086.42, a=(4.10537, -0.331101, 3.40482, 494.15, -24.2444), lam = 1e-11
err = 717.823, a=(4.25593, -0.339248, 3.40224, 493.248, -25.0837), lam = 1e-12
err = 482.563, a=(4.37647, -0.345234, 3.40072, 492.673, -25.7561), lam = 1e-13
err = 332.278, a=(4.47295, -0.349724, 3.3998, 492.292, -26.2945), lam = 1e-14
err = 236.222, a=(4.55013, -0.353147, 3.39923, 492.031, -26.7256), lam = 1e-15
err = 174.801, a=(4.61188, -0.355787, 3.39888, 491.848, -27.0707), lam = 1e-16
err = 135.516, a=(4.66126, -0.357842, 3.39865, 491.718, -27.3471), lam = 1e-17
err = 110.383, a=(4.70075, -0.359454, 3.39852, 491.624, -27.5683), lam = 1e-18
err = 94.3008, a=(4.73234, -0.360724, 3.39843, 491.556, -27.7453), lam = 1e-19
err = 84.0091, a=(4.75759, -0.361731, 3.39839, 491.506, -27.8871), lam = 1e-20
err = 77.4223, a=(4.77778, -0.362531, 3.39836, 491.469, -28.0006), lam = 1e-21
err = 73.2062, a=(4.79393, -0.363169, 3.39836, 491.442, -28.0914), lam = 1e-22
err = 70.5075, a=(4.80683, -0.363678, 3.39836, 491.422, -28.1641), lam = 1e-23
err = 68.7799, a=(4.81715, -0.364085, 3.39836, 491.408, -28.2223), lam = 1e-24
err = 67.6739, a=(4.8254, -0.364412, 3.39837, 491.397, -28.2689), lam = 1e-25
err = 66.9659, a=(4.832, -0.364674, 3.39838, 491.389, -28.3062), lam = 1e-26
err = 66.5126, a=(4.83727, -0.364884, 3.3984, 491.383, -28.336), lam = 1e-27
err = 66.2223, a=(4.84149, -0.365053, 3.39841, 491.379, -28.3599), lam = 1e-28
err = 66.0365, a=(4.84486, -0.365189, 3.39842, 491.376, -28.379), lam = 1e-29
err = 65.9175, a=(4.84755, -0.365298, 3.39843, 491.374, -28.3944), lam = 1e-30
err = 65.8414, a=(4.84971, -0.365386, 3.39844, 491.372, -28.4066), lam = 1e-31
Best-fit parameters: M = 4.84971, e = -0.365386, omega = 0.256843, tP = 491.372, v0 = -28.4066
t = np.linspace(tobs[0], tobs[-1], 1000)
plt.plot(t, rv(t,P, a), 'C0')
plt.plot(tobs, vobs, 'C1.')
plt.errorbar(tobs, vobs, eobs, fmt='none', ecolor='C1')
plt.ylabel(r'$v$')
plt.xlabel(r'$t$')
plt.show()
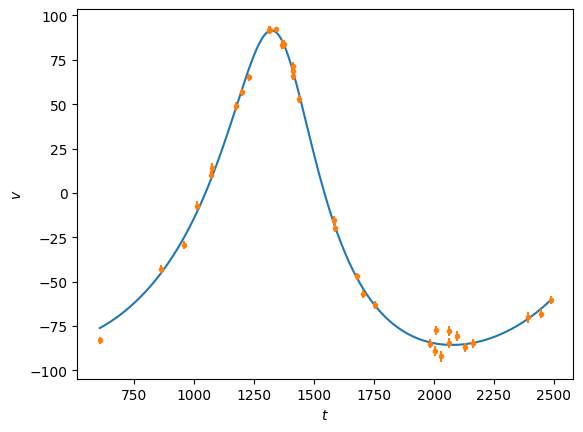
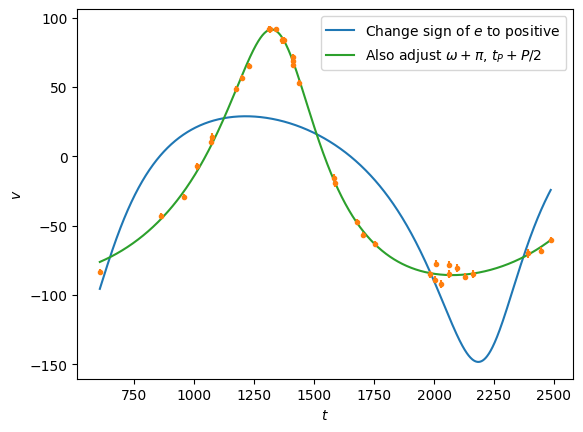
Note that the eccentricity from the fit comes out negative, this is because of a degeneracy between \(e\) and the angles \(\omega_P\) and \(t_P\). By adjusting these, we can fix the eccentricity to be positive:
t = np.linspace(tobs[0], tobs[-1], 1000)
# Change the sign of eccentricity
a2 = np.copy(a)
a2[1] = -a[1]
plt.plot(t, rv(t,P, a2), 'C0', label=r'Change sign of $e$ to positive')
a3 = np.copy(a)
a3[1] = -a[1]
a3[3] = (a[3] + P/2) % P
a3[2] = (a[2] + np.pi) % (2*np.pi)
plt.plot(t, rv(t,P, a3), 'C2', label=r'Also adjust $\omega+\pi$, $t_P+P/2$')
plt.plot(tobs, vobs, 'C1.')
plt.errorbar(tobs, vobs, eobs, fmt='none', ecolor='C1')
plt.ylabel(r'$v$')
plt.legend()
plt.xlabel(r'$t$')
plt.show()
# Calculate the covariance matrix
v, A = rv_model(tobs, P, a3)
A[:, 0] = A[:, 0] / eobs
A[:, 1] = A[:, 1] / eobs
A[:, 2] = A[:, 2] / eobs
A[:, 3] = A[:, 3] / eobs
A[:, 4] = A[:, 4] / eobs
C = np.linalg.inv(A.T@A)
To compare with MCMC, let’s regenerate the samples (I copied the code directly from the exercise here: https://andrewcumming.github.io/phys512/metropolis_solutions.html)
seed = 56123
rng = np.random.default_rng(seed)
def f(x, tobs, vobs, eobs):
chisq = np.sum(((vobs-rv(tobs, P, x))/eobs)**2)
return -chisq/2
# Observations
# These are for HD145675 from Butler et al. 2003
tobs, vobs, eobs = np.loadtxt('rvs.txt', unpack=True)
# Number of samples to generate
N = 10**5
x = np.zeros((N, 5))
# initial guess
# P, mp, e, omega, tp, v0
P = 1724
x[0] = [1.0, 0.0, 0.0, 0.0, 0.0]
# and the widths for the jumps
widths = (0.03, 0.03, 0.03, 3.0, 1.0)
count = 0
for i in range(N-1):
# Proposal
ii = np.random.randint(0, 5)
x_try = np.copy(x[i])
x_try[ii] += rng.normal(scale = widths[ii])
#x_try[2] = (x_try[2]) % 1 # keep e between zero and 1
# Accept the move or stay where we are
u = rng.uniform()
if u <= np.exp(f(x_try, tobs,vobs,eobs) - f(x[i], tobs,vobs,eobs)):
x[i+1] = np.copy(x_try)
count = count + 1
else:
x[i+1] = np.copy(x[i])
print("Acceptance fraction = %g" % (count/N,))
# Reject the burn in phase
x = x[int(0.3*N):]
# Move the angles to within 0 to 2pi
x[:,2] = x[:,2] % (2*np.pi)
x[:,3] = x[:,3] % P
Acceptance fraction = 0.39794
# Plot to make sure everything looks ok
titles = (r'$M/MJ$', r'$e$', r'$\omega$', r'$t_P$', r'$v_0$')
for i, title in enumerate(titles):
plt.subplot(2,3,i+1)
plt.title(title)
plt.hist(x[:, i], density=True, bins=30)
plt.tight_layout()
plt.show()
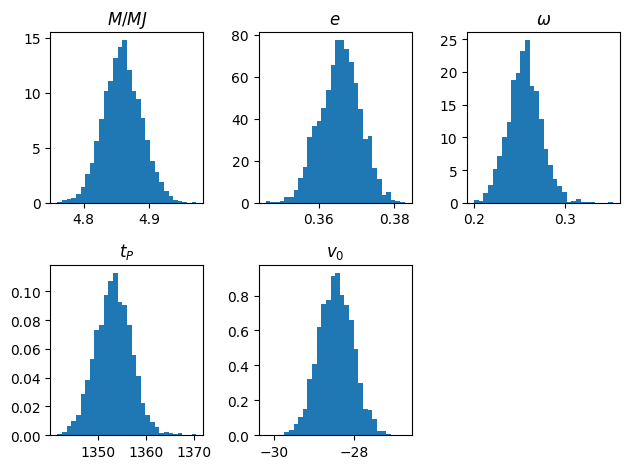
# compare with the least squares results
print('%12s %12s %12s %12s %12s %12s' % ('parameter', 'MCMC', 'LS', 'MCMC var', 'LS var', 'frac diff'))
for i in range(5):
print("%12s %12lg %12lg %12lg %12lg %12lg" %
(titles[i], np.mean(x[:,i]), a3[i], np.var(x[:,i]), C[i,i],
np.divide(np.var(x[:,i]) - C[i,i], np.var(x[:,i]) + C[i,i])))
parameter MCMC LS MCMC var LS var frac diff
$M/MJ$ 4.85904 4.84971 0.000844139 0.000848462 -0.00255406
$e$ 0.365621 0.365386 2.84255e-05 2.87864e-05 -0.0063075
$\omega$ 0.255935 0.256843 0.000334086 0.000377849 -0.061471
$t_P$ 1353.21 1353.37 14.2288 16.2268 -0.0656025
$v_0$ -28.4507 -28.4066 0.184243 0.183562 0.00184951
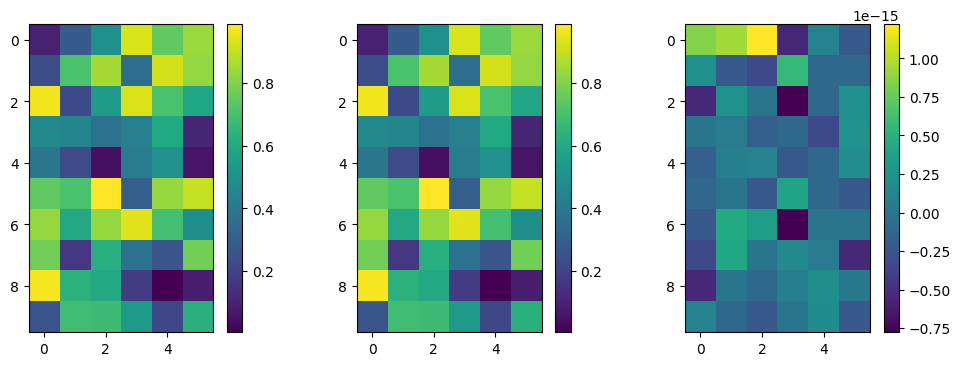
Good agreement! Let’s also check the full covariance matrix:
C_mcmc = np.cov(x.T)
with np.printoptions(precision=3, suppress=False):
print(C,"\n\n",C_mcmc)
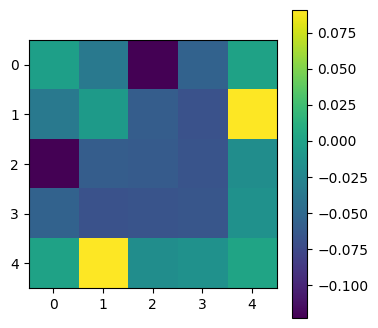
plot_matrices([np.divide(C_mcmc-C, C_mcmc+C)])
[[ 8.485e-04 -6.036e-05 -5.155e-05 -1.316e-02 -4.161e-03]
[-6.036e-05 2.879e-05 -8.196e-06 -2.972e-03 -3.427e-04]
[-5.155e-05 -8.196e-06 3.778e-04 7.299e-02 4.870e-04]
[-1.316e-02 -2.972e-03 7.299e-02 1.623e+01 6.866e-02]
[-4.161e-03 -3.427e-04 4.870e-04 6.866e-02 1.836e-01]]
[[ 8.442e-04 -5.610e-05 -4.027e-05 -1.178e-02 -4.166e-03]
[-5.610e-05 2.843e-05 -7.275e-06 -2.589e-03 -4.113e-04]
[-4.027e-05 -7.275e-06 3.341e-04 6.376e-02 4.694e-04]
[-1.178e-02 -2.589e-03 6.376e-02 1.423e+01 6.671e-02]
[-4.166e-03 -4.113e-04 4.694e-04 6.671e-02 1.842e-01]]
<Figure size 640x480 with 0 Axes>