Homework 5 solutions#
import numpy as np
import matplotlib.pyplot as plt
import scipy.integrate
import time
Adaptive Runge Kutta integration of orbits#
def rk4_step(t, dt, x, derivs):
f = derivs(t, x)
f1 = derivs(t + dt/2, x + f*dt/2)
f2 = derivs(t + dt/2, x + f1*dt/2)
f3 = derivs(t+dt, x + f2*dt)
return x + dt*(f + 2*f1 + 2*f2 + f3)/6
def rk4_driver(dt, t_end, x_start, derivs, tol=1e-6):
x = np.array([x_start])
tvec = np.array([])
t = 0.0
tvec = np.append(tvec,t)
while t < t_end:
while True:
x1 = rk4_step(t, dt, x[-1], derivs)
x2 = rk4_step(t, dt/2, x[-1], derivs)
x3 = rk4_step(t, dt/2, x2, derivs)
err = max(abs((x3-x1)))
if err < tol:
t = t + dt
tvec = np.append(tvec,t)
dt = dt * 2
# check to see whether this step size would
# take us beyond t=t_end and if so adjust accordingly
if t + dt > t_end:
dt = t_end - t
# store the more accurate (2-step) result
x = np.row_stack((x, x3))
break
else:
dt = dt / 2
return tvec, x
def derivs_orbit(t, x):
rr = (x[0]**2 + x[1]**2)**0.5
vxdot = -x[0]/rr**3
vydot = -x[1]/rr**3
xdot = x[2]
ydot = x[3]
return np.array((xdot,ydot,vxdot,vydot))
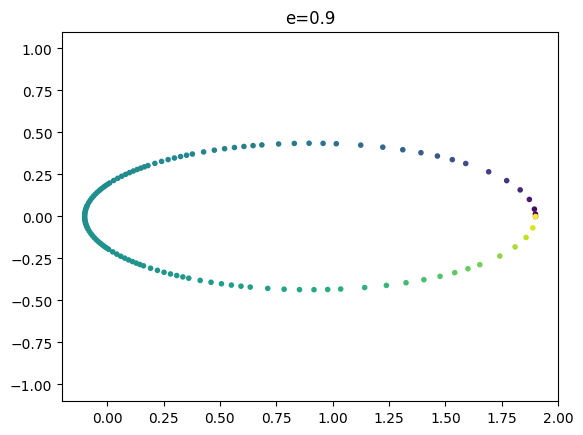
r = 1.9 # starting radius = 1+e
nsteps = 100
dt = 2 * np.pi / (nsteps-1)
t = np.arange(nsteps)*dt
x_start = np.array((r,0,0,np.sqrt((2/r)-1)))
t4, x4 = rk4_driver(dt, 2*np.pi, x_start, derivs_orbit, tol=1e-6)
print('number of integration points = %d' % (len(x4,)))
print('integrated to t/2pi=%lg' % (t4[-1]/(2*np.pi)))
print('start to end differences:', x4[0]-x4[-1])
v = np.sqrt(x4[:,2]**2 + x4[:,3]**2)
print('vmin=%g, vmax=%lg' % (min(v), max(v)))
r = np.sqrt(x4[:,0]**2 + x4[:,1]**2)
print('rmin=%g, rmax=%lg' % (min(r), max(r)))
e = (max(r)-min(r))/2
print('eccentricity of the orbit = %lg' % (e,))
cols = 255 * t4/max(t4)
x = x4[:,0]
y = x4[:,1]
plt.scatter(x, y, marker = '.', c=cols.astype(int).tolist())
plt.title(r'e=%.3lg' % (e,))
plt.xlim((-(1-e)-0.1,1+e+0.1))
plt.ylim((-1.1,1.1))
plt.show()
number of integration points = 125
integrated to t/2pi=1
start to end differences: [ 9.81260632e-07 -5.15549565e-07 6.00980559e-07 -1.14780523e-07]
vmin=0.229416, vmax=4.3589
rmin=0.1, rmax=1.9
eccentricity of the orbit = 0.9
Method of lines#
def calculate_A(n, h, banded = True):
# calculate the matrix A = 1+Ch
# represented in banded form
b = (1+2*h)*np.ones(n)
a = -h*np.ones(n)
c = -h*np.ones(n)
c[-1] = 0.0
a[0] = 0.0
# boundary conditions: the temperatures at the ends are fixed
# so we need to adjust the matrix to give T_end^(n+1) = T_end^n
# x=0
b[0] = 1.0
a[1] = 0.0
# x=1
b[-1] = 1.0
c[-2] = 0.0
if banded:
AA = np.row_stack((a,b,c))
else:
AA = np.diag(b, k=0) + np.diag(a[1:], k=1) + np.diag(c[:-1], k=-1)
return AA
t0 = time.time()
n = 1001
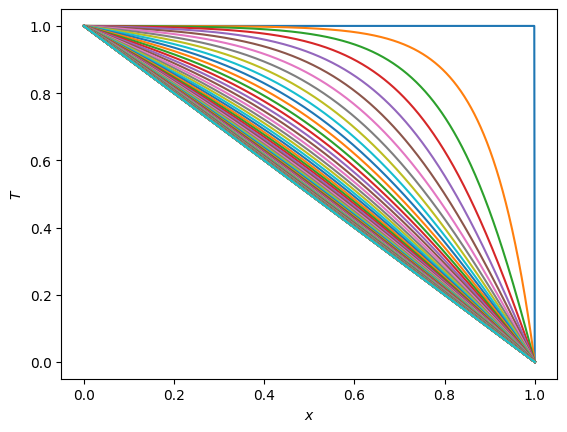
x = np.linspace(0, 1, n)
# Initial profile has T=1 everywhere except T=0 at the outer boundary
T = np.ones(n)
T[-1] = 0.0
plt.plot(x, T)
plt.xlabel(r'$x$')
plt.ylabel(r'$T$')
dx = x[1]-x[0]
dt = 1e-2
h = dt/dx**2
nsteps = 100
AA = calculate_A(n, h)
# store the gradient at the top
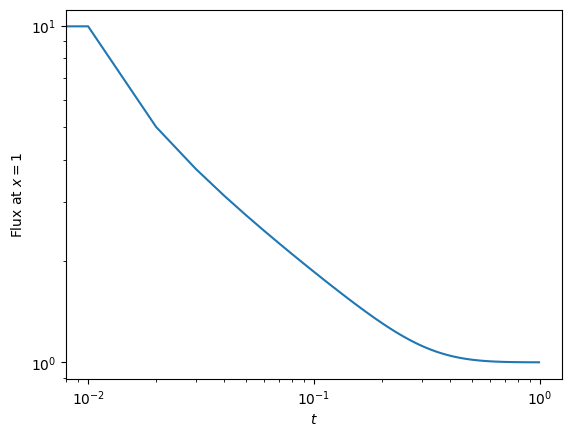
F = np.zeros(nsteps)
F[0] = -(T[-1]-T[-2])
for i in range(1,nsteps):
T = scipy.linalg.solve_banded((1,1), AA, T)
F[i] = -(T[-1]-T[-2])/dx
plt.plot(x, T)
print('Time taken = ', time.time()-t0)
plt.show()
plt.clf()
plt.plot(np.arange(nsteps)*dt, F)
plt.xlabel(r'$t$')
plt.ylabel(r'Flux at $x=1$')
plt.yscale('log')
plt.xscale('log')
plt.show()
Time taken = 0.045166015625
# Repeat but now calculate the full matrix inverse instead of using tridiagonal solver
t0 = time.time()
# Initial profile has T=1 everywhere except T=0 at the outer boundary
T = np.ones(n)
T[-1] = 0.0
plt.plot(x, T)
plt.xlabel(r'$x$')
plt.ylabel(r'$T$')
AA = calculate_A(n, h, banded=False)
# store the gradient at the top
F = np.zeros(nsteps)
F[0] = -(T[-1]-T[-2])
for i in range(1,nsteps):
Ainv = np.linalg.inv(AA)
T = Ainv@T
F[i] = -(T[-1]-T[-2])/dx
plt.plot(x, T)
print('Time taken = ', time.time()-t0)
plt.show()
plt.clf()
plt.plot(np.arange(nsteps)*dt, F)
plt.xlabel(r'$t$')
plt.ylabel(r'Flux at $x=1$')
plt.yscale('log')
plt.xscale('log')
plt.show()
Time taken = 3.045142650604248


